MetaboLights
MetaboLights is a data repository specific to metabolomics studies 1. You can submit data from NMR, GC-MS, LC-MS, and MALDI amongst others.
The repository is maintained and curated by the European Bioinformatics Institure (EMBL-EBI) meaning that the data it holds is well-formatted and integrated with a number of other standardized databases and ontologies (ways of describing methods, data and metadata). This “future-proofs” the data you store, making it not only open-access but also more findable and re-useable, as well as meaning it can be integrated with other -omics data if required.
⚠️ You’ve worked hard to get this data, so make sure it gets stored somewhere that other people can make use of it!
Submission
MetaboLights has various stages of submission, validation and then curation by experts to make sure your submission has all the relevant metadata needed to recreate your analysis. Following curation, there is a review process and finally your data can be added to the repository and made available on MetaboLights.
Because of the curation process, there can be some lag between submission and your data being available so you should submit in as much time as possible. However, as soon as you’ve submitted, you will have a reference that can be linked to any publication.
Firstly you will need to create an account by clicking on “Submit to MetaboLights” on the MetaboLights landing page.
You will then be able to watch a video tutorial guide on using the submission portal.
Hints and tips
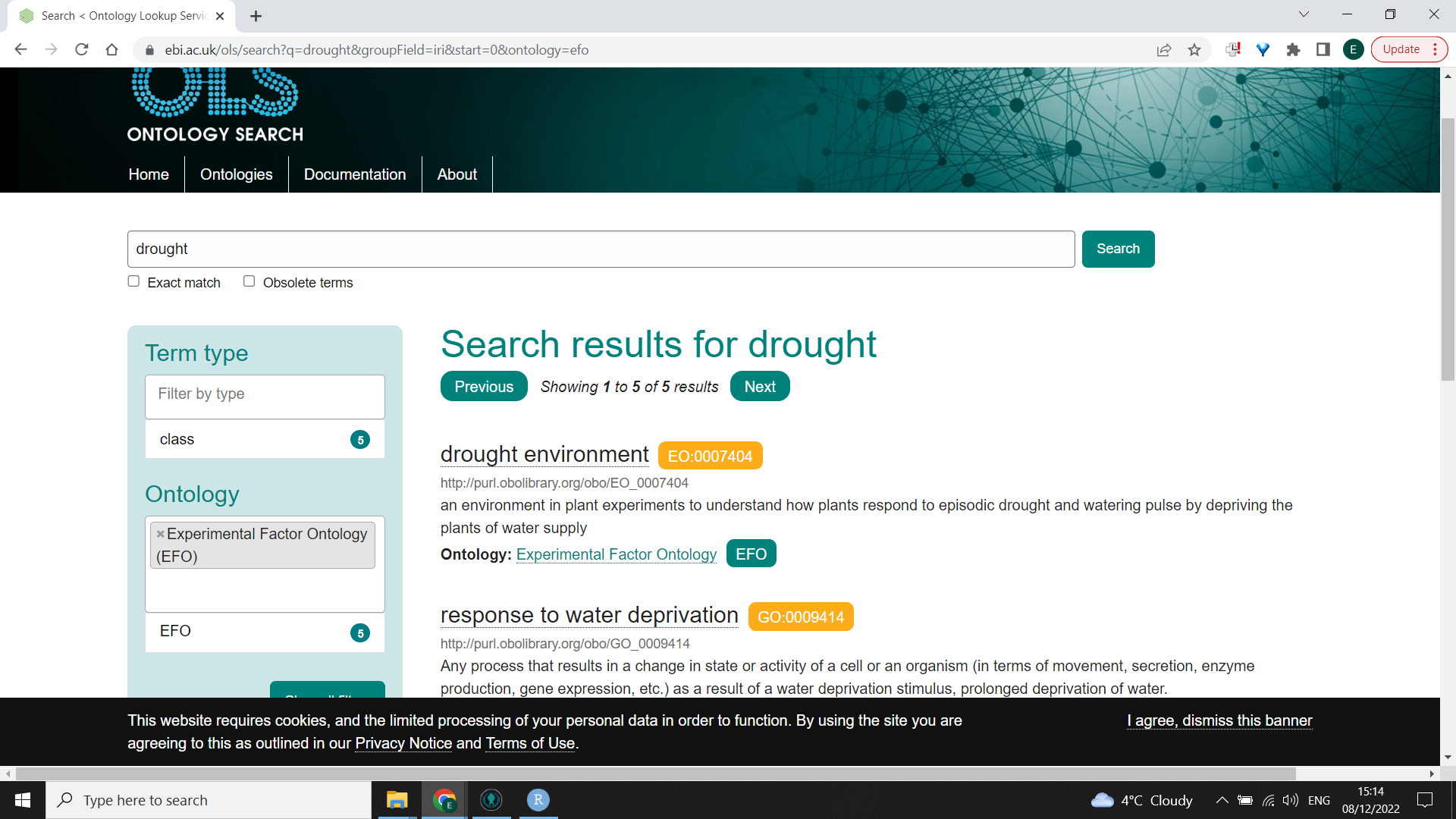
- You will need to understand the principle of shared ontologies to be able to label your variables and experimental set up. EMBL have a useful ontology lookup tool. In the image below, I have searched for “drought” and then filtered by “experimental factor ontology” because I wanted to know how to describe one of my variables (an imposed drought treatmnent). I can then use the code “EO:0007404” to label that variable in my submission to MetaboLights.
-
You will need to report your methodology, including all the parameters and settings used for your mass spectrometry. These can be found in the “_HEADER.txt” file in your original .RAW folders from MassLynx. You will also need to report your processing and analytical workflow and all the tools you have used (this may help you collate these details).
-
Use a good internet connection for upload of files and try to be patient.
-
You will upload .raw files to the RAW_FILES folder and .mzML files to the DERIVED_FILES folder. Make sure the names of the files exactly match those you enter in the appropriate columns of the “Assays” table.
-
If you get stuck, email metabolights-help@ebi.ac.uk and someone will respond to you (be nice, they’re real people!).
-
Kenneth Haug, Keeva Cochrane, Venkata Chandrasekhar Nainala, Mark Williams, Jiakang Chang, Kalai Vanii Jayaseelan, Claire O’Donovan (2020). MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Research 48(D1):D440–D444 https://doi.org/10.1093/nar/gkz1019 ↩︎