KEGG
Using KEGG to help with annotation
Once you have a list of your putative IDs from METLIN, cross reference these with KEGG COMPOUND. This will give you a lot of information about the biological roles of any putative compounds.
💡 KEGG COMPOUND IDs begin with a C followed by 5 or 6 digits e.g. L-lysine is C00047
KEGG PATHWAY is a very useful tool for outlining the metabolic processes of a wide range of organisms and you can discount some compounds that are not likely to be produced by your organism.
⚠️ be careful with discounting compounds in this way if you have a multi organism system or one where compounds amy have been introduced from another source than our study organism. For example:
- 🍄 roots of arbuscular mycorrhizal plants will contain both plant and fungal compounds
- 🦠 root exudates will contain compounds that bacteria biosynthesize but plants don’t
- 🌱 plant compounds can be used as antimicrobials and medical (human or vetinary) drugs
- 🚜 field samples may contain pesticides and contaminants
Using KEGG PATHWAY
Open KEGG PATHWAY > Change Pathway Type > Use Ctrl + F and enter the name of your organism > click on the three letter code to the left of your organism This will code all your KEGG pathways to highlight those that are relevant to your organism.
💡 If there is not a map for your particular organism it is worth doing more investigation into your potential identification to see what kind of compound it is and whether that might feasibly be present in your organism.
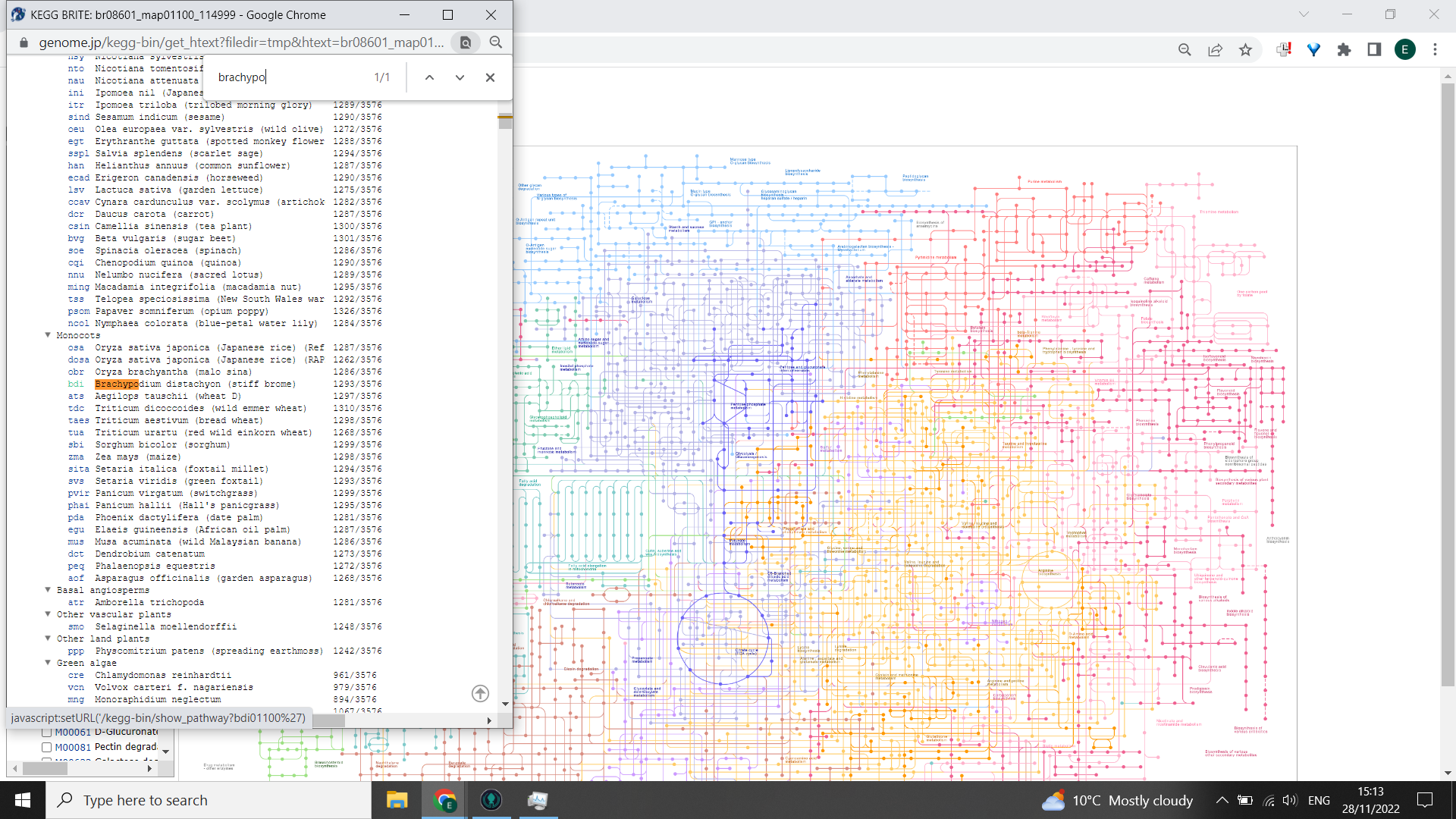
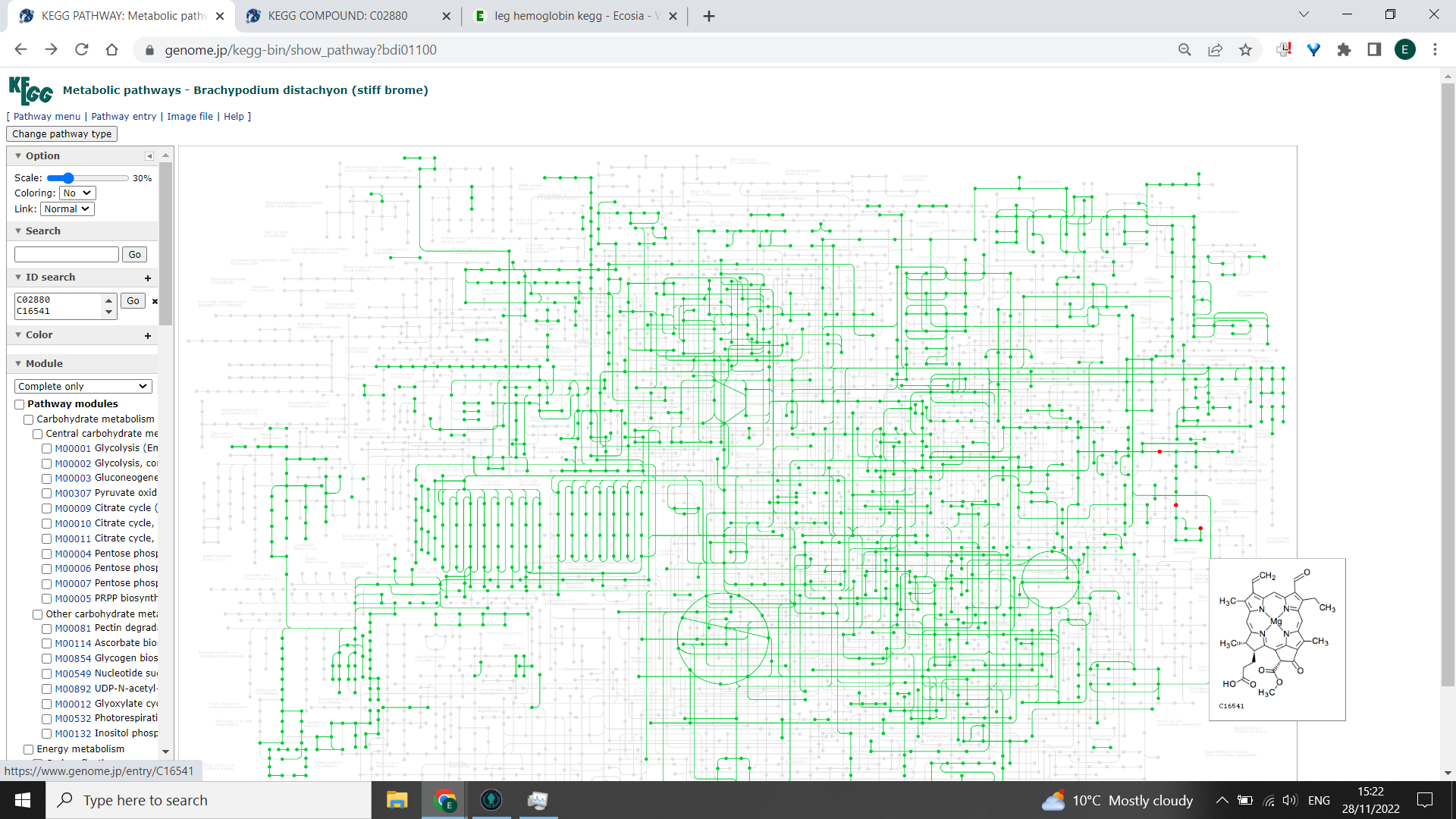
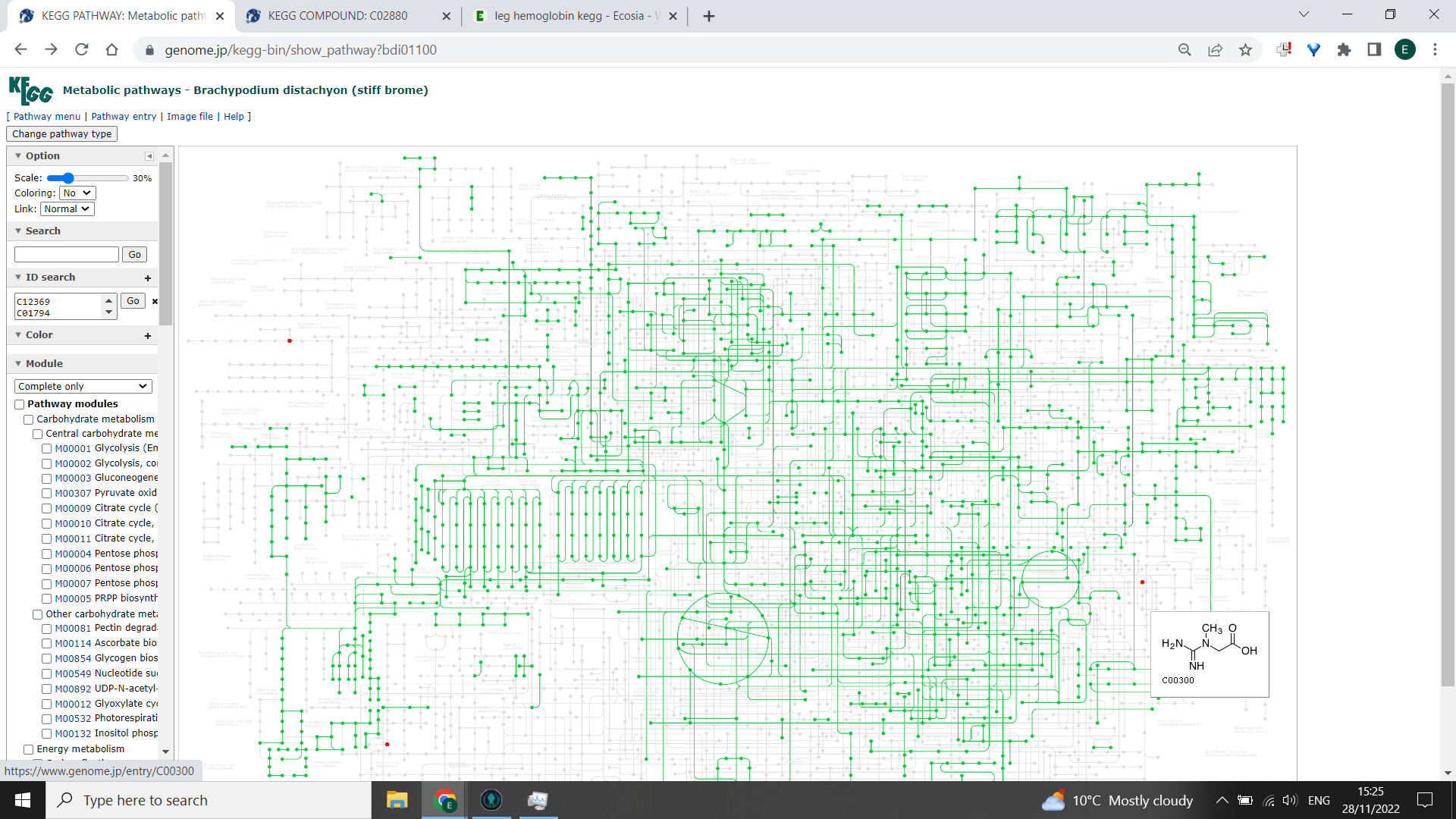
Now, in the box to the left hand side that says “ID search”, paste your list of KEGG IDS and click “Go”. You should get some nodes highlighted red on the KEGG PATHWAY. If you hover the cursor over each point, it will tell you the KEGG ID and name - any that are highlighted but are in the greyed out portion of the pathway are unlikely to be produced by your study organism.
⚠️ You should further cross reference with other metabolite databases to discern whether this is a sensible identification.
Three red nodes highlighted are likely to be found in this organism, forming part of their global metabolism:
Three red nodes highlighted here are NOT likely to be found in this organism, as they do not form part of it’s global metabolism map. In this example, they may be kingdom specific:
Other databases to consult
- MassBank
- PubChem
- E.coli Metabolite Database
- MetaCyc (includes organism-specific databases which can help narrow down annotation)
- Metabolomics workbench has a further list of useful databases
If there are published metabolomics studies of your organism or a closely related one, then you can compare your mz (or mz/RT combinations) to theirs by searching their data in a repository such as MetaboLights.
⚠️ In all cases using this method, your MSI annotation level will be 3 as all these methods go no further than comparison with the literature and repositories.
For an MSI annotation level 1 or 2, you must do further targeted MS, tandem MS or NMR, or analyse the specific compounds you have highlighted in this untargeted approach.