Processing LCMS
Pre-processing LC-ESI-MS data with XCMS Online
XCMS online allows you to run the XCMS R package remotely, which is very useful if you have a lot of data or a complex experimental structure. The goal here is to produce a peak table that you can take forward to other analysis tools. We don’t recommend using XCMS for statistics because it doesn’t allow for normalisation and scaling, nor for detailed annotation as it is unlikely to be specific to your study organism.
⚠️ SIGN UP to XCMS ASAP - it can take a few days for your password to become active.
Go to XCMS online and log in or register (free but must provide an email address and agree to the terms - should be cited in any publication).
Uploading your data
Select “Stored Datasets” > “Add”
Type your dataset name and select “Save” which saves the name only, not the files.
Select “Browse” to choose your .mzml files from ONE group or condition of your experiment. Do not close the window while files are uploading. Once all files have a green tick next to them select “Save Dataset & Proceed”.
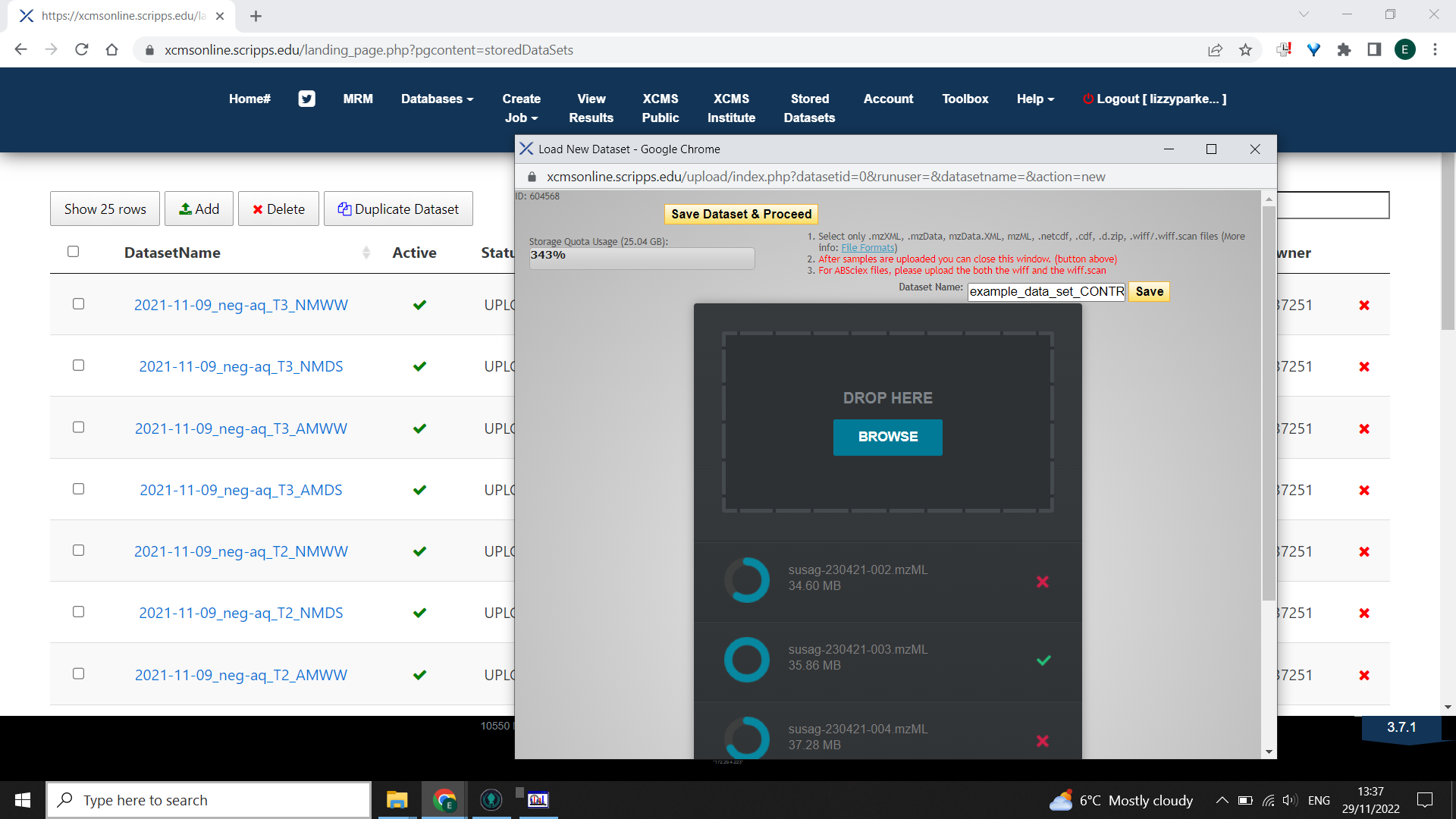
Do this for all your data groups. So if you have Case vs. Control, you will have two “data sets” to upload.
💡 XCMS suggests an upload limit but we find you can surpass this
Setting up your processing job
Option A: To compare two groups choose a Pairwise comparison
From the homepage “Create job” > “Pairwise”
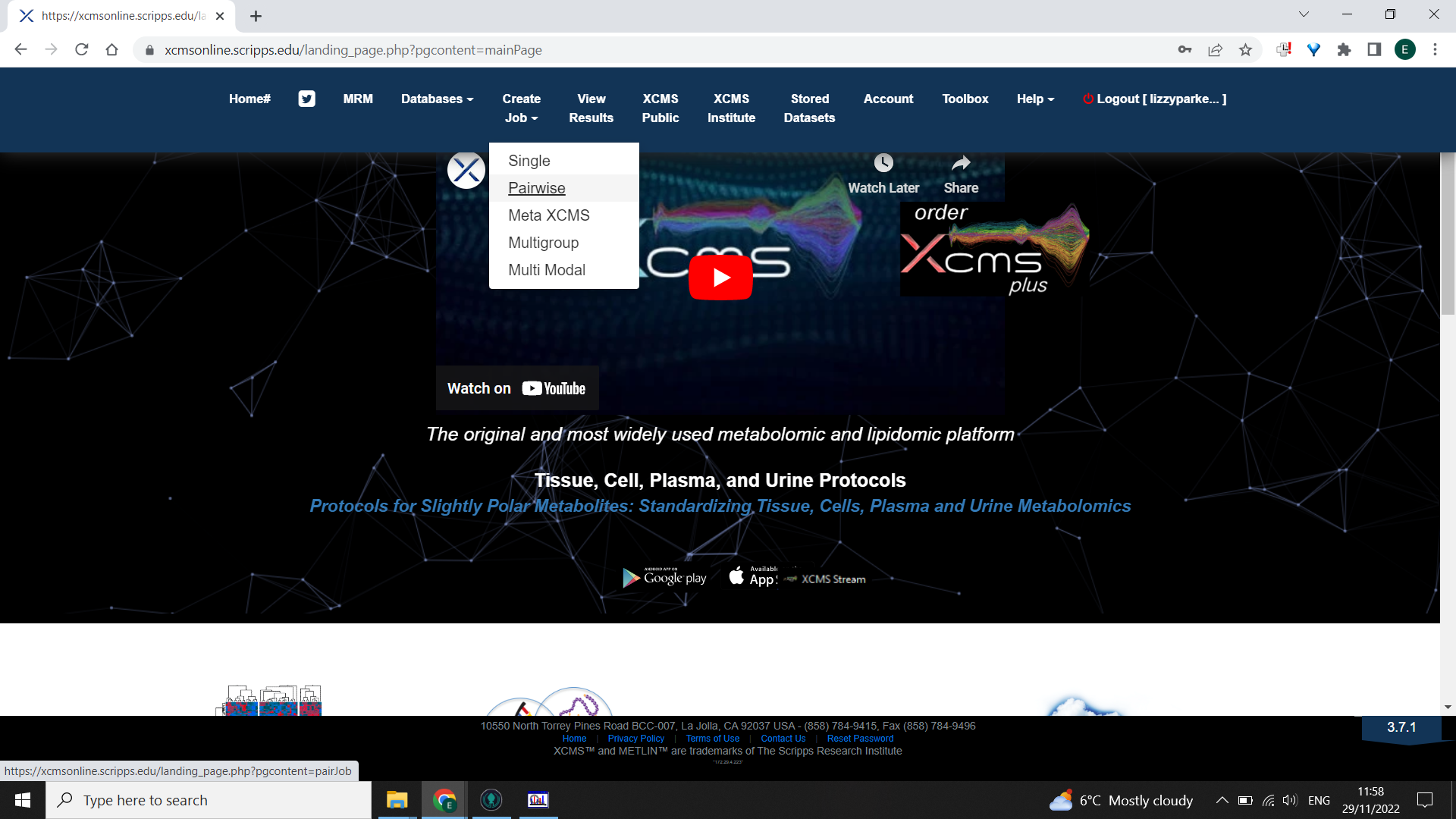
Select the first dataset to load in “Select dataset” > “Next” Select the second dataset of the comparison with “Select dataset” > “Next”
Option B: To compare more than two groups choose a Multigroup comparison
From the homepage “Create job” > “Multi-group” Use “Select Dataset” to choose the datasets (groups) to compare in your analysis and then close the pop-up window once you have made your selection.
Click “Next”.
Choosing parameters
Choose the most appropriate Parameters based on the kind of mass spectrometry that was done. An example is shown below for an LCMS dataset.
📖 For more information on choosing your parameters Forsberg et al. 2018 1 is very helpful. The IPO package in R is very useful for gathering information about the data. Usually this is done with a small subset of your data - e.g one file from each group but can be slow and require a lot of computing power.
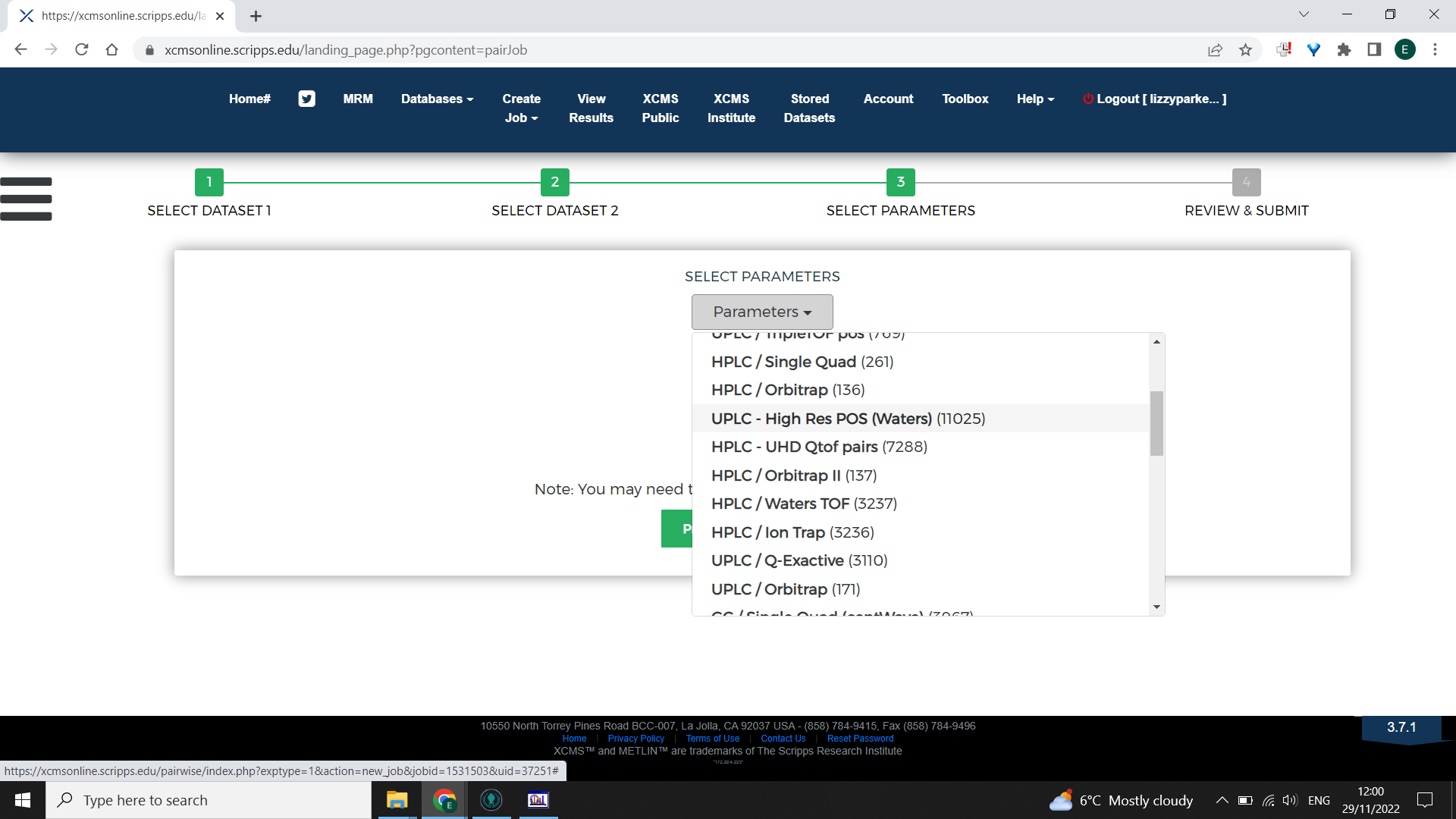
It is possible to tailor the parameters to be optimal for your dataset. To do this, begin with selecting the closest appropriate XCMS online parameters then select “View/Edit” > “Create new”
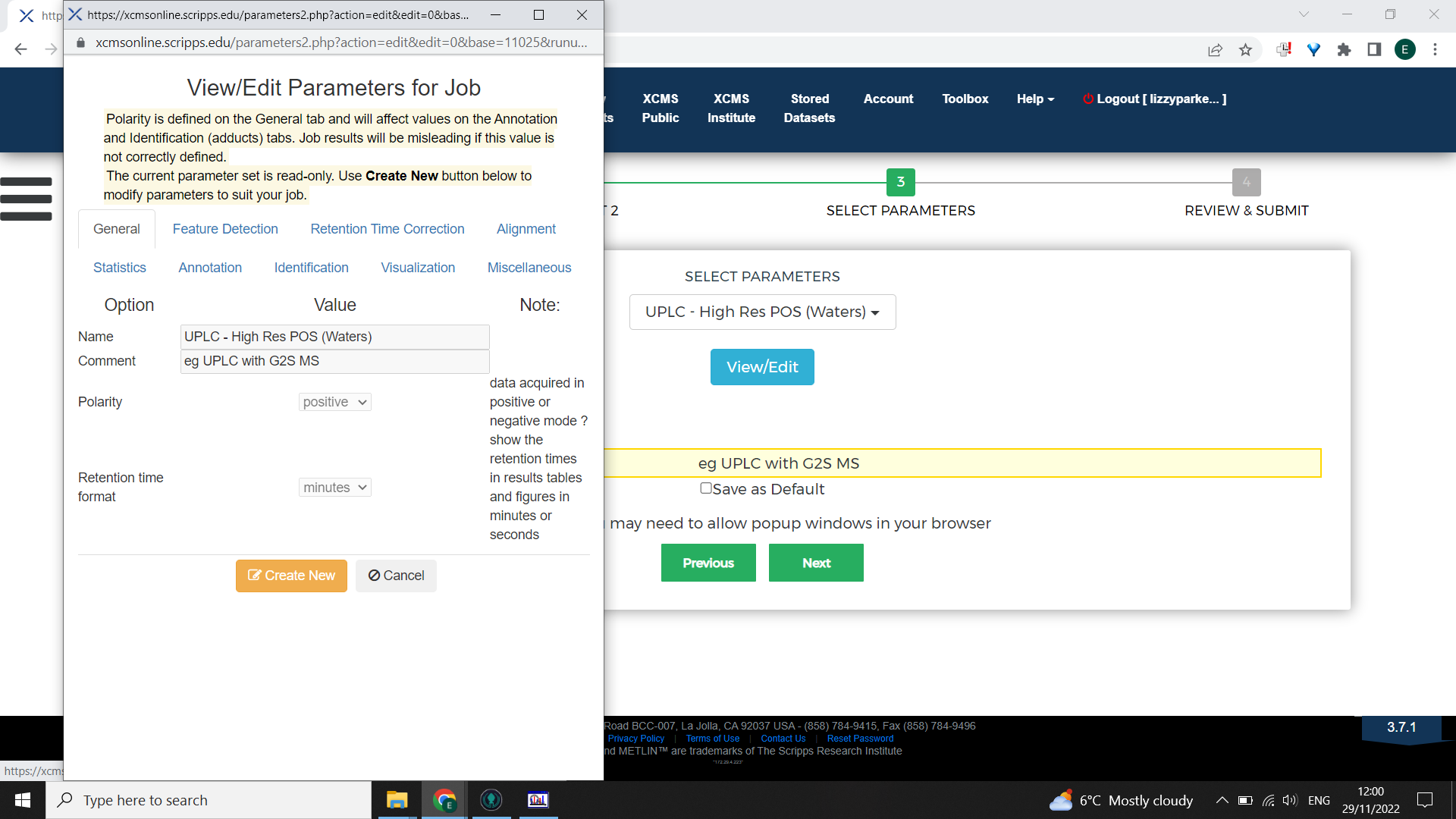
Work through the tabs inputting the information from IPO or Forsberg et al 2018. Then rename the parameter set something meaningful and save.
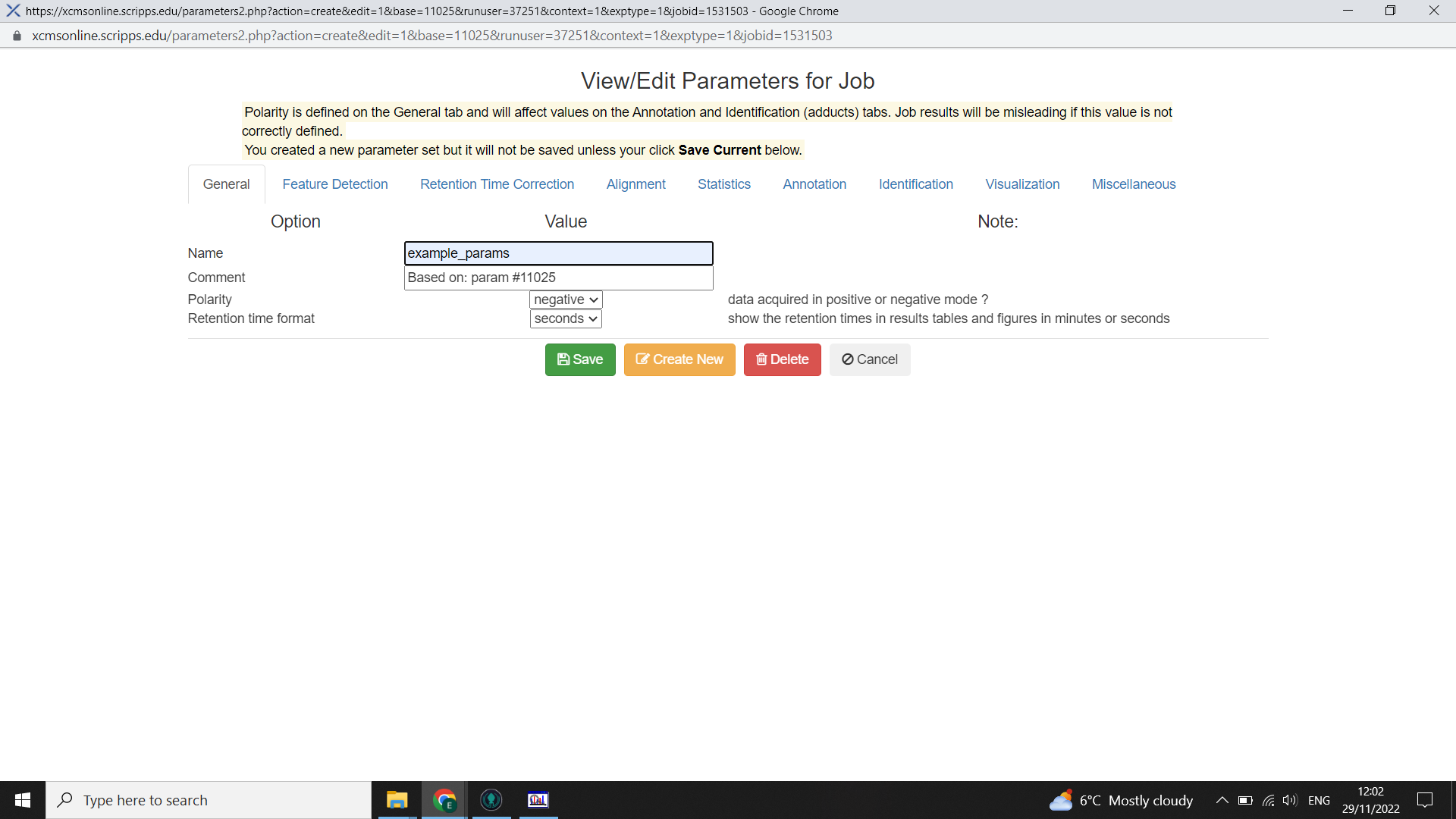
Submitting your processing job
Select “Submit” job, you will receive email notifications about the progress of your job e.g. whether it has failed or when it has finished.
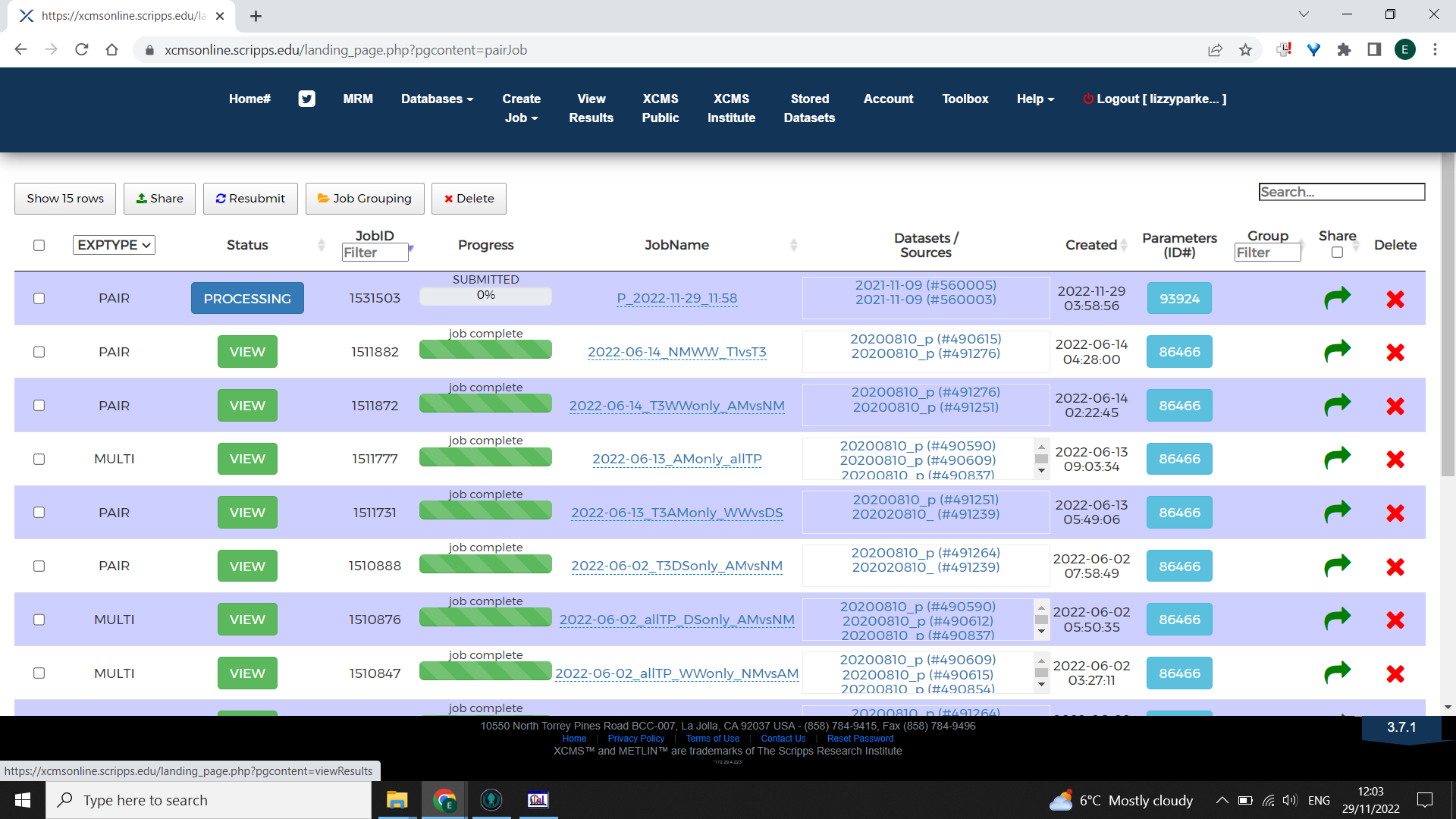
When complete, select the “View Results” tab from the homepage “View” the appropriate job and click “Download Results” in the top right corner.
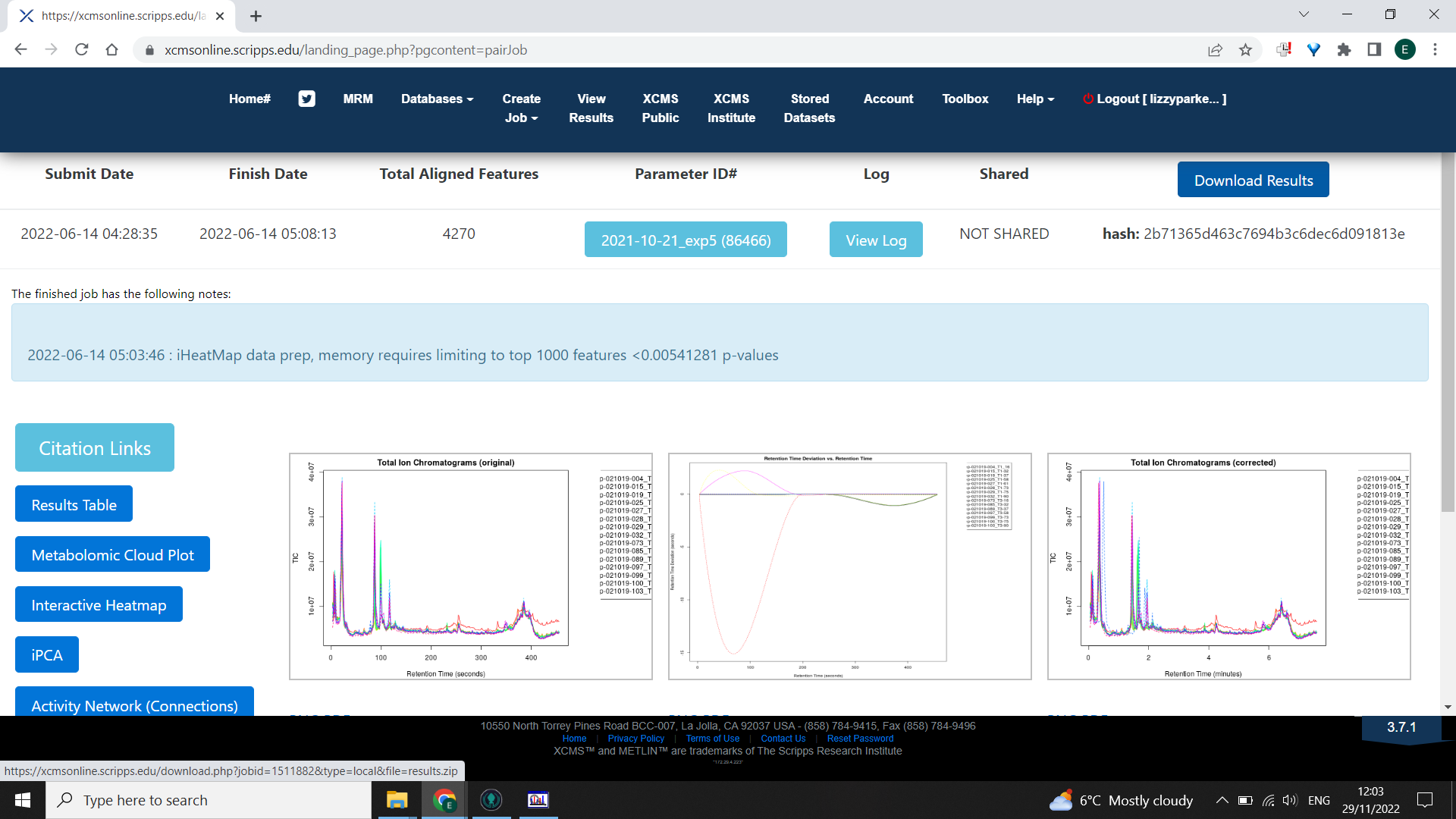
⚠️ You can only have one job running at a time (but you can close the website whilst it’s running - it’s working remotely).
-
Forsberg, E., Huan, T., Rinehart, D. et al. (2018). Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat Protoc 13, 633–651 DOI: https://doi.org/10.1038/nprot.2017.151 ↩︎