METLIN
Using METLIN to help with annotation
METLIN has a really good search feature with which we can define a lot of search criteria to make our lives a bit easier.
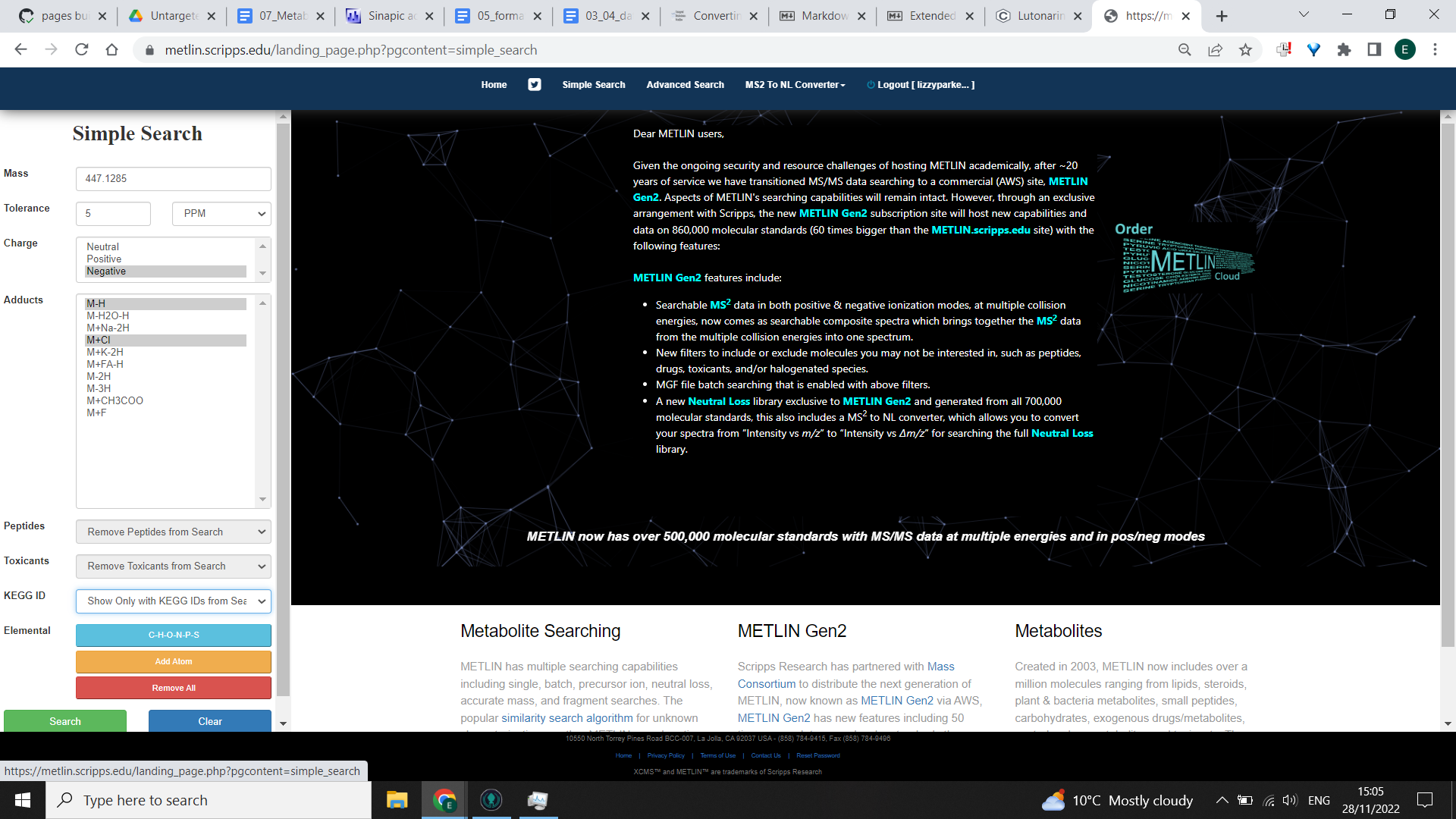
Firstly log in and select “Simple search”
Enter your mass of interest (ideally to 4 or 5 decimal points).
Tolerance: Choose the tolerance (we would recommend using 5ppm to comply with MSI guidance).
Charge: positive or negative depending on which mode your data was run in.
Adducts: go with the defaults (most common adducts) or if you have used certain MS techniques, certain adducts may be more likely
For biological samples I’d recommend choosing to see only those samples that have KEGG IDs associated with them (from the KEGG ID drop down list). This is likely to remove many synthetic chemicals that are unlikely to appear in organism metabolisms naturally and reduces the number of options you need to go through.
Create a spreadsheet of these results. You will need to report:
- detected mass (what you put into your search)
- accurate mass
- chemical formula
- adduct formed
- ppm (this gives an idea of how closely matched to expected monoisotopic mass your detected mass was)
- compound name
- METLIN ID and KEGG ID
- For reporting on MetaboLights, ChEBI identifier is particularly useful though they do provide a lookup tool so this can be added at a later stage
⚠️ METLIN currently locks you out temporarily if it “detects automated searching” (if you search compounds too quickly one after another). It will let you back in after around 20mins