MSConvert MALDI
Convert MALDI data to open format
Open MSConvert
⚠️ If you are using a remote drive (e.g. google drive or onedrive) with a slow connection it is worth doing your conversion in small batches so you can check the file size as you go - if you are seeing big discrepencies you need to go back and check the sizes of your original files. Some files may need re-converting if your connection times out during the process.
File: “Browse” > Select .RAW folders to convert
Output Directory: Specify an output folder using “Browse”
Options: Output File > mzML
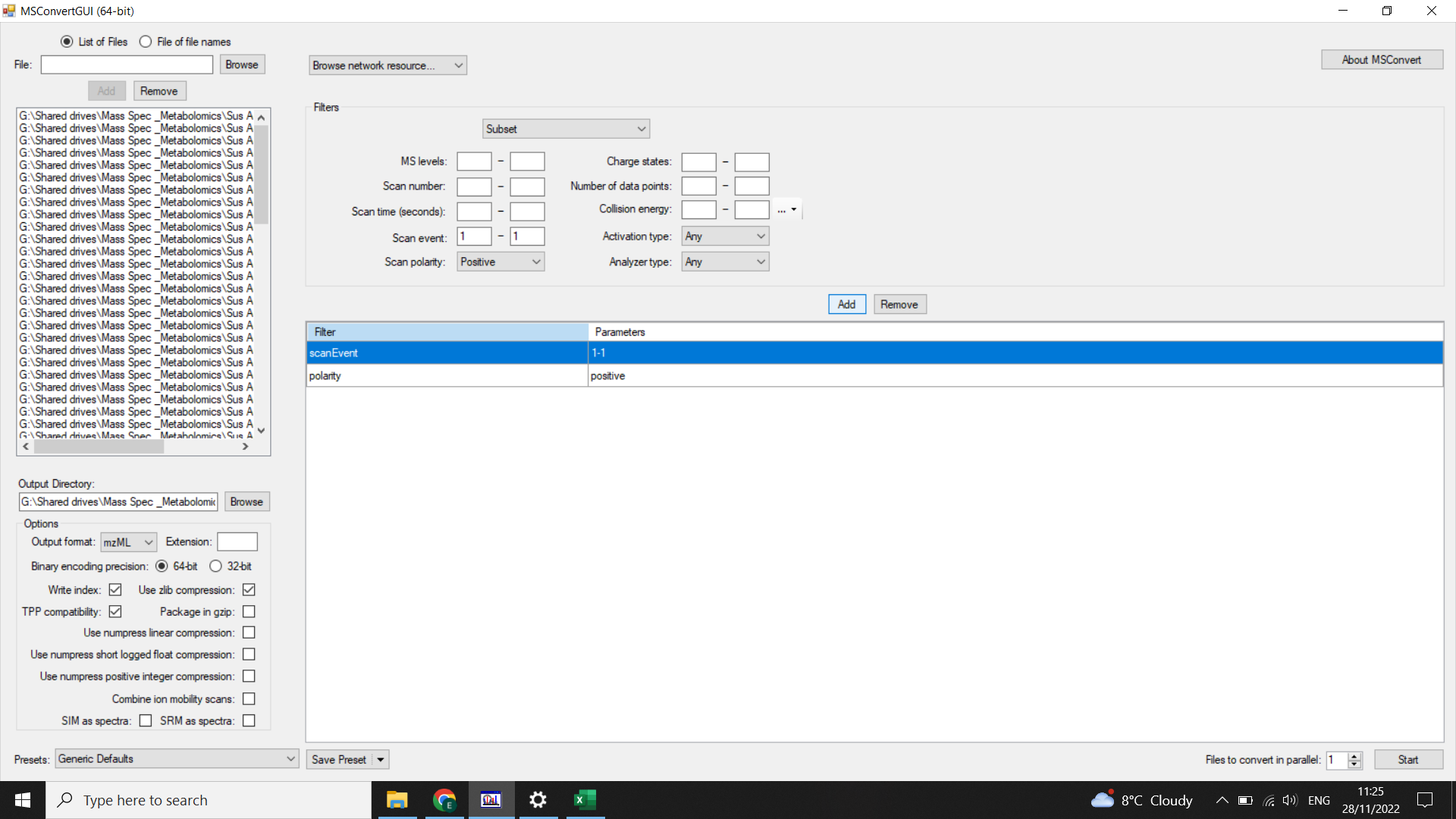
Filters: select “subset” (from drop-down menu) > “scanNumber” = 2-119 (we want to remove the first scan, use SeeMS to check the number of the last scan and adjust this value), and select “polarity = positive” or “polarity = negative” depending on your MS run (and click “add”)
Select “Start” (bottom right)
N.B. for MALDI data we do not perform PeakPicking during conversion because we need to maintain the profile shape of the data (rather than centroiding)
Check the size of .mzML files - should all be similar. Reconvert any that look small. If you consistently have some that are much smaller, it is worth checking whether these ran properly or whether there were any issues copying or converting the files (such as intermittent internet connection).
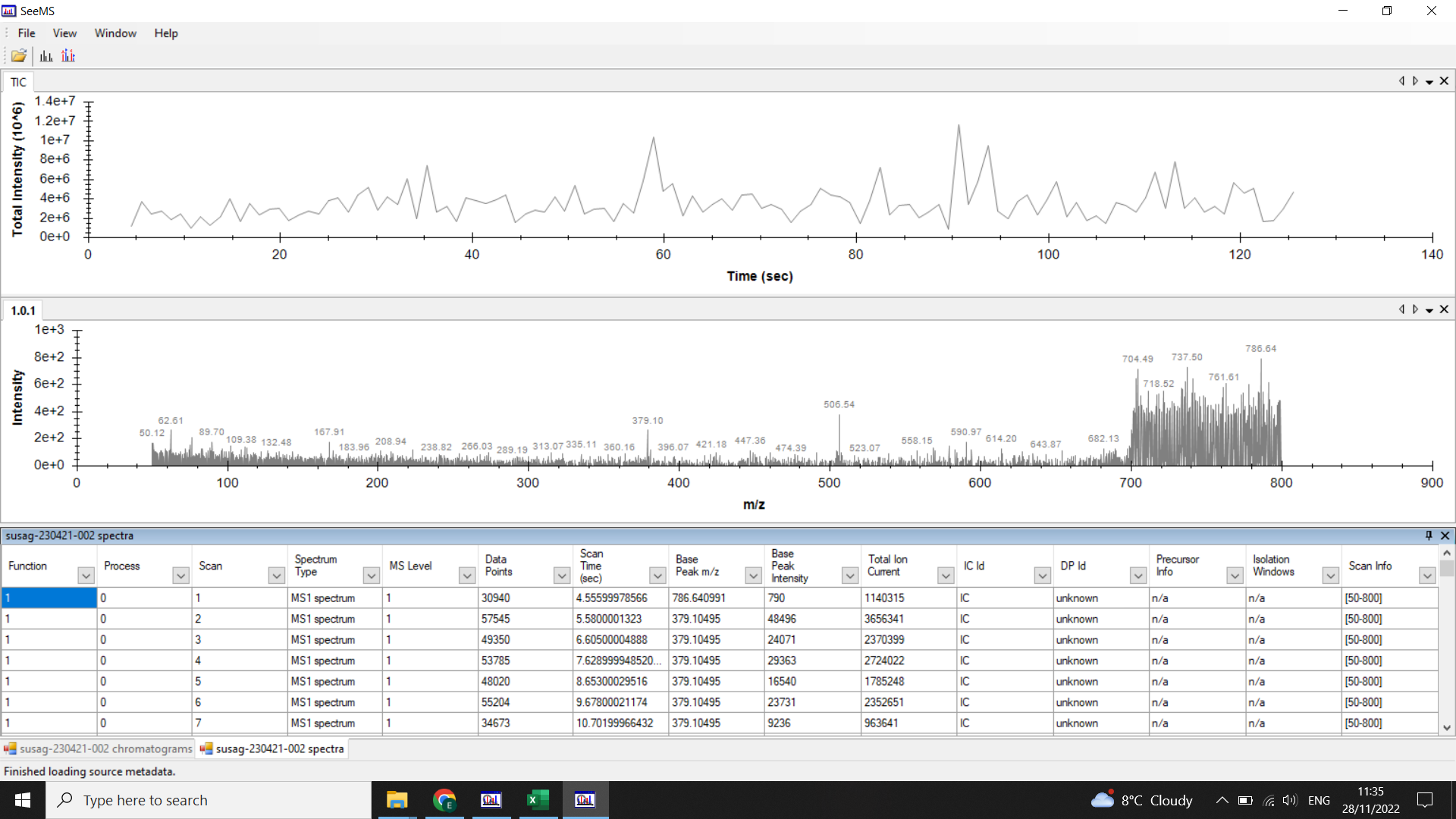
⚠️ If you have any big gaps or empty tables when you look at your chromatograms/ spectra in SeeMS, then you should go back and redo the conversion.